BNTL-Multi-Sources
Bayesian Network Transfer Learning (BN-TL) algorithm to re-use of source model through multiple data sources
This package is in private, and please contact author to get package.
1. Introduction
The goal of this project is to increase the re-use of computable biomedical knowledge in probabilistic formalisms. In the field of transfer learning, which is a sub-area of machine learning, these differences between settings are referred to as heterogeneity. Unlike traditional machine learning, transfer learning explicitly distinguishes between a source setting, from which we develop a model M that we would like to re-use, and a target setting, for which we use or adapt M, often because the target setting has insufficient data to develop a model de novo. Researchers distinguish transfer learning algorithms by what kinds of heterogeneous transfer learning scenarios (or simply heterogeneous scenarios) they can handle.
This project created the Bayesian Network Transfer Learning (BN-TL) algorithm to re-use of source model, such as influenza, learned from electronic medical record (EMR) data to predict the target data set. (for more info see: Transfer Learning For Bayesian Case Detection Systems)
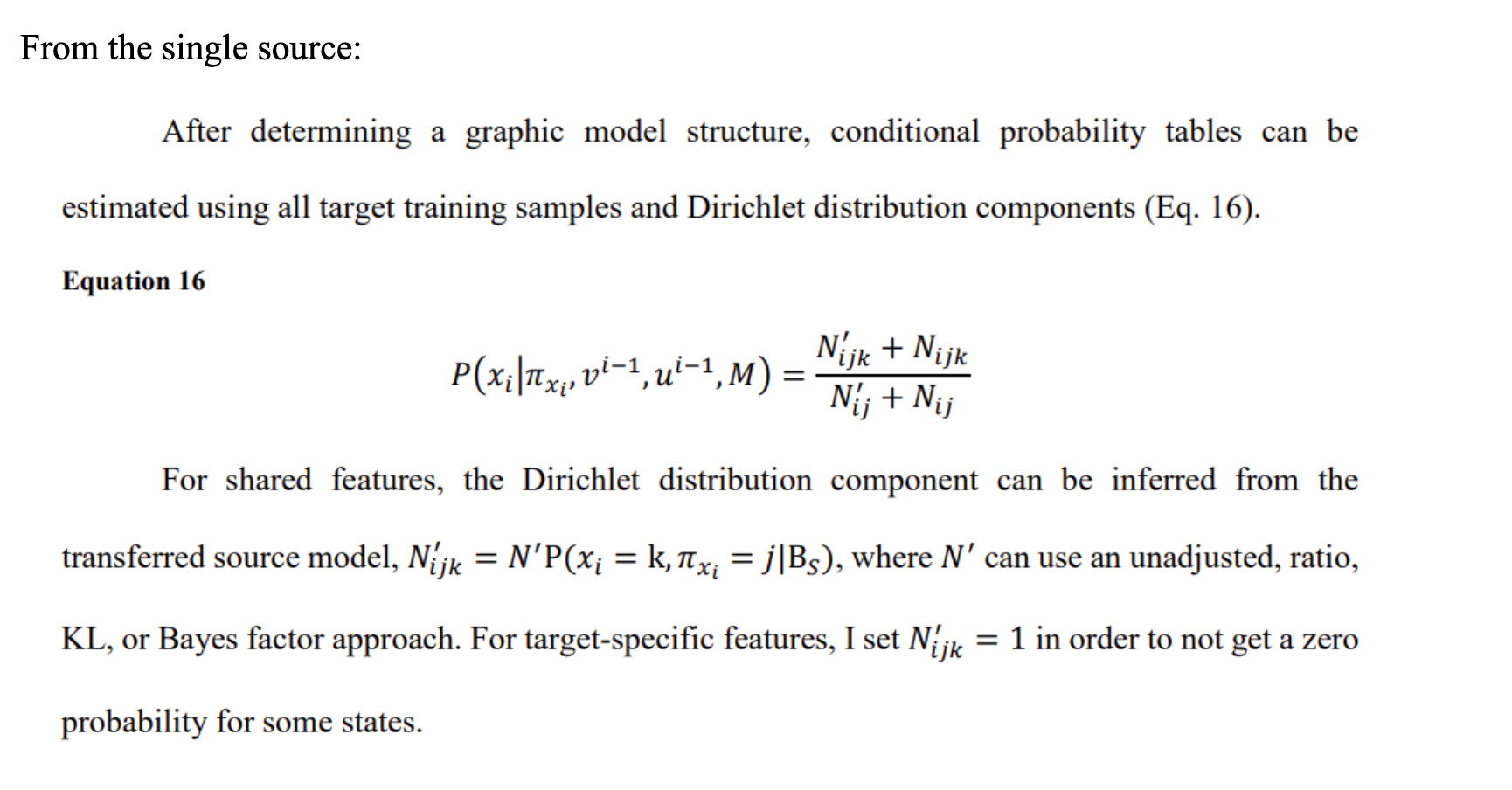
2. Quick Start
If you have not install JAVA JDK 18 or 1.8 yet, please install first
2.1 Set Up
-
Git clone https://github.com/AI-In-Population-Health-Lab/BNTL_multiSource.gitor download as zip file - download your get 6-month smile key –> put into
smile_license folder
2.1.1 CLI - Quick Start
cd BNTL current direcotry-
java -jar BNTL_MultiSource.jar==> this will show the brief instruction of the project. java -Djava.library.path=./lib -jar BNTL_MultiSource.jar ./smile_license/License.java ./no_KL FLU-SLC2AC-0809-transfer20090118-IG001-NB unadjust ratio off
2.1.2 IntelliJ IDEA
- Open BNTL project
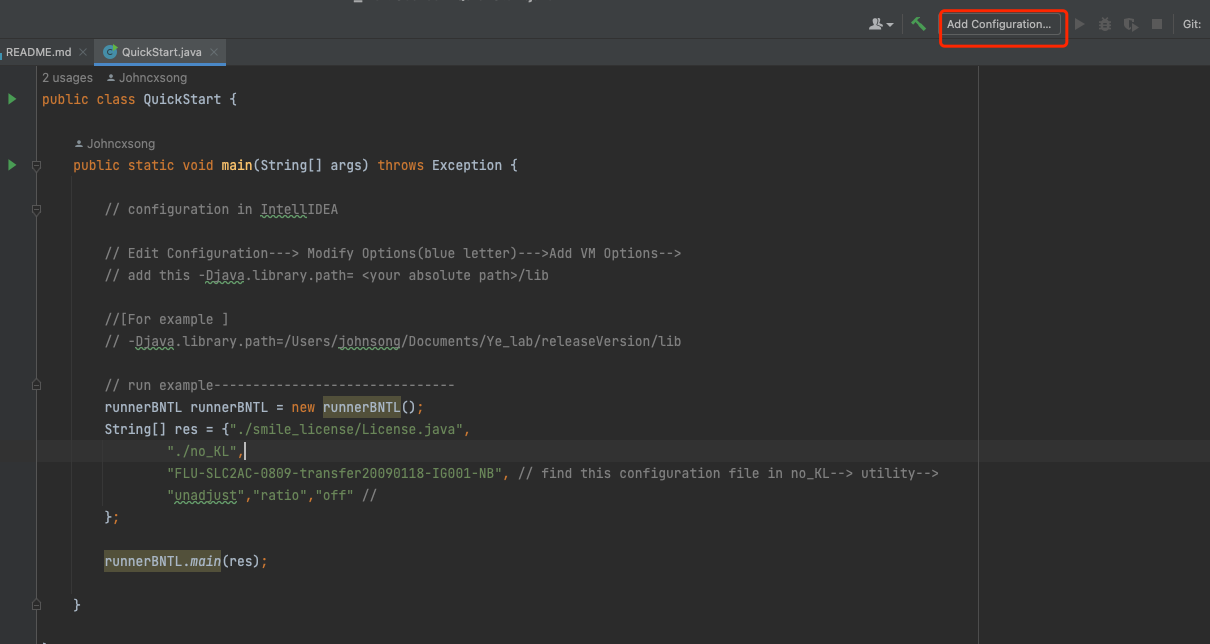
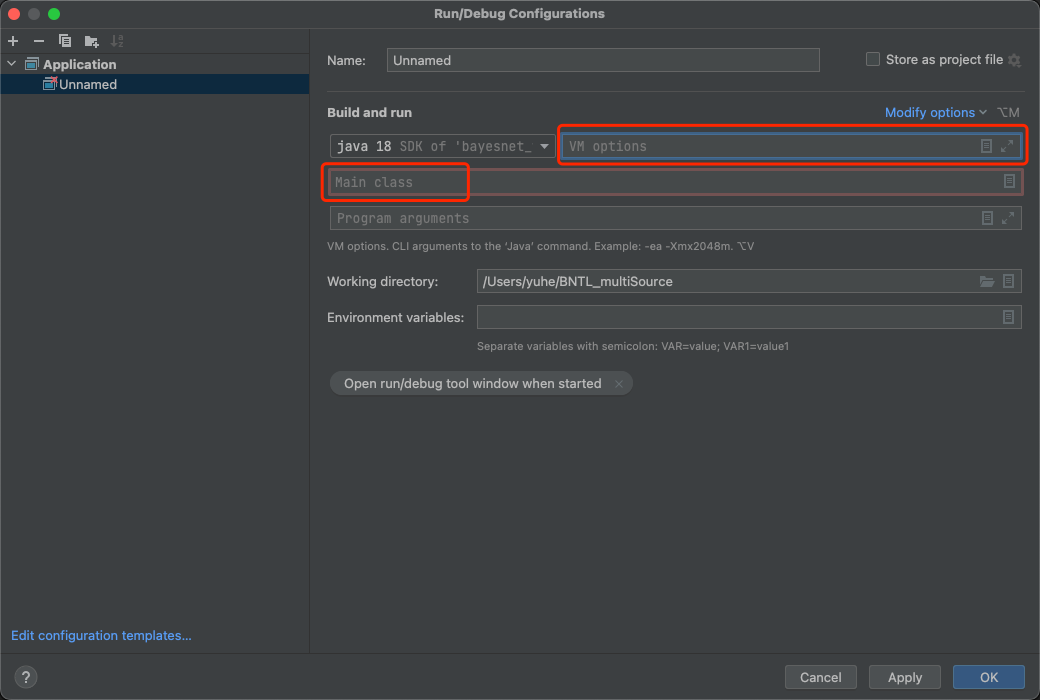
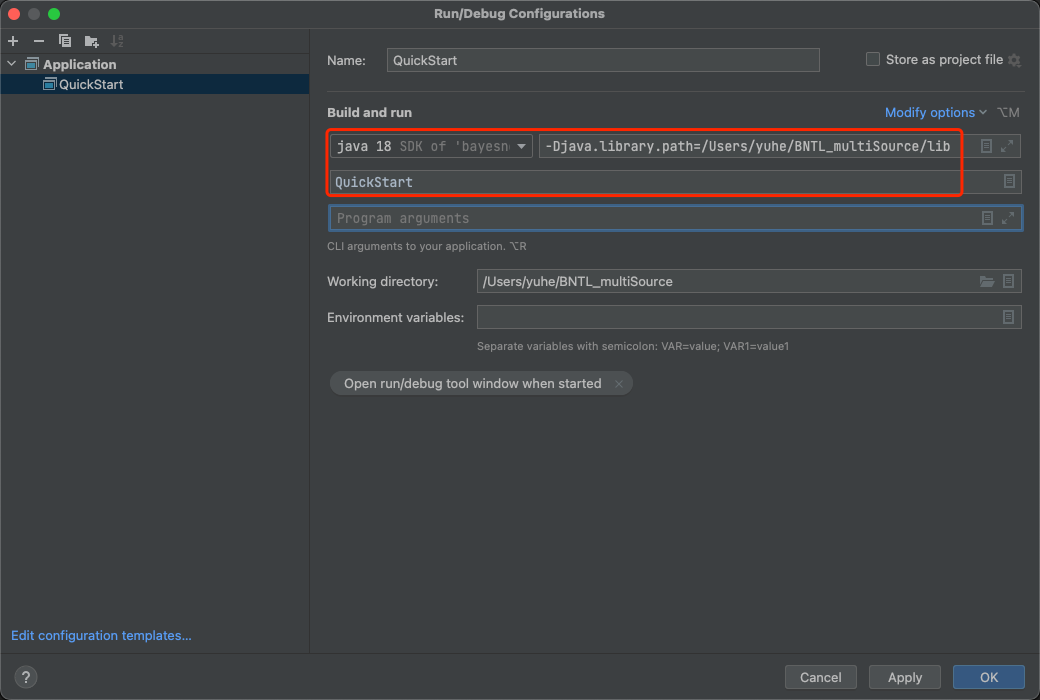
- Src folder —> QuickStart.java
- Check QuickStart.java comments for configuration to add smile package into library.
- Run
3. Run your own models
3.1 Prepare input folder
- data must be in .arff format.
- model must be in .bif, .xml, .xdsl format.
3.1.1 structure of input folder
folder_name
utility
your configurationFile.txt
sourceData
sourceData.arff
sourceModel
source_learn_model.bif (you could put multi-models)
targetData
target_test_data.arff
target_train_data.arff
targetModel
result
intermediate folder-->to store result.
log
3.1.2 parameters for configurationFile.txt
-----Source Part-----
sourceDataLoc=
sourceModelLoc=
sourceDataName=source_data.arff
sourceLearnedModelName= 1.bif,2.bif,...
sourceDataSize=1661,1661....
-----Target Part-----
targetNodeName=diagnosis
targetTrainDataSize=21
targetTestDataName=test_data.arff
targetTestDataSize=344
targetModelLoc=path--->targetModel/
targetDataName=train_data.arff
targetDataLoc=path-->targetData/
-------feature-------
modelLearningApproach=NB
featureSelectionApproach=IG001
-------Path------
temporaryFileName=temp.xml
experimentFileLoc= path to folder
utilityLoc=path-->/utility/
resultProbName=prob.csv
resultAUCName=auc.csv
logName=configuration_file.txt
resultLoc=path--->result/
logLoc=path-->log/
hashCodeName=hashCode.xml
------------KL----------(if you want to perform KL section)
trueKL=1.8224292702088067
trueBFTable=hypoxemia,1.0E-9;reported_fever,1.0E-9;influenza_lab_positive,1.0E-9;age_group,1.0E-9;diagnosis,1.0E-9;unspecified_cough,3.2417270528642096E-6;nasal_swab_order,1.0E-9;
KL_targetData_learnedSourceModel=1.2568181979662876
nodeKLTable_targetData_learnedSourceModel=diagnosis,0.0045124560875614075;unspecified_cough,0.09813077790883455;reported_fever,0.4261776773290663;age_group,0.01770522273709965;hypoxemia,0.47951291320078326;nasal_swab_order,0.114442540587127;
KL_targetData_trueSourceModel=1.660745666413443
nodeKLTable_targetData_trueSourceModel=diagnosis,0.03595232239858527;age_group,0.0281376732895748;nasal_swab_order,0.05359926700974576;unspecified_cough,0.019446531674810627;reported_fever,0.4892853435516517;influenza_lab_positive,0.09896658196551227;hypoxemia,0.8059805217479895;
- run unadjust & ratio performance, you need to have Source Part, Target Part, Feature Part, and Path part.
- Run KL options, you need to have KL Part that includes in configuration file.
- For multi-source processing,
sourceLearnedModelName= 1.bif,2.bif,... sourceDataSize=1661,1661...
4. Weighted & Multi-Sources
4.1 Weighted (unadjusted & Ratio)
Searching model process is led by a score. The score is the sum of log marginal likelihood and log structure prior
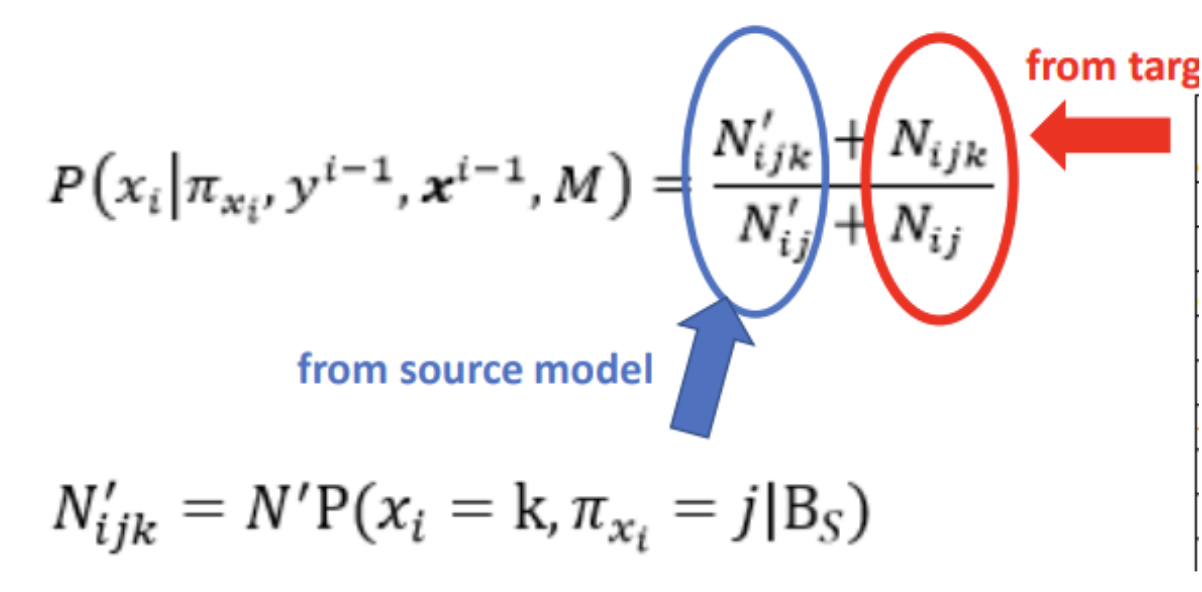
4.1.1 Unadjusted Approach
Use sample size of the source training data.
4.1.2 Ratio Approach
Use sample size of the target training data.
Number of target instances / number of source training instances
Weight the source sample size based on the similarity between the target distribution and the source distribution. If the source and the target distributions are very different, it would be better to assign a lower equivalent sample size for a transferred source model.

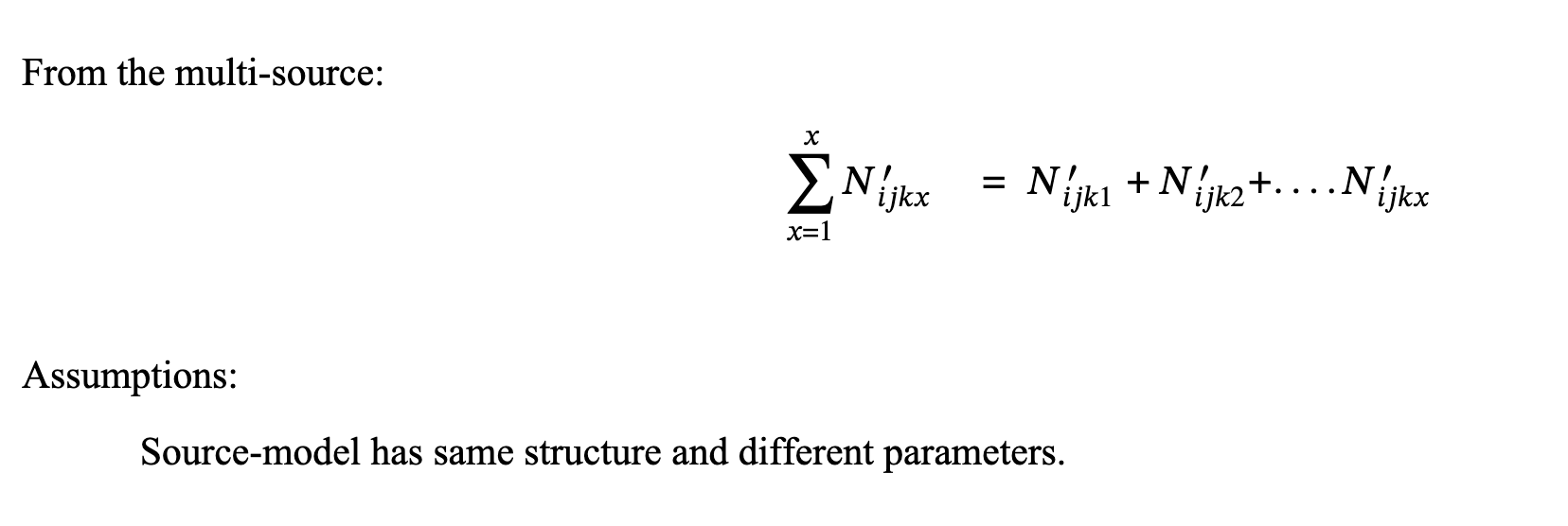
4.2 Multi-Sources
5. Resources
- Weka package It provides data format structure, the Bayes Network Structure, classifier, evaluation methods.
- Jsmile packageIt is encoding the Bayes Network for depth of probability calculation.